1: DLS and SEC methods
2: M&M protein purification
3: Label free measurement of the effect of HNK on Sirt
4: M7M Chemicals and Reagents.
I tried to put those protocols together, SM please read through #1, 2 and see if there is any factual inaccuracies.
SM: 1 and 2 are accurate.
==
AU: 6-7-2017: Regarding method section of template document-Are we including Sirt3 purchased from Enzo life Sciences as indicated in "chemicals and reagents"?
Thank you
RC: Unless we report any results with it (even in context of controls mentioned), no.
--
AU: 5-31-2017: Solubility measurements-
Solubility measurements.docx
Same file has been uploaded in dropbox (Dropbox\PMC-AT PLIN\PMCAT Fitting)
RC: PMC-AT fitting is not the right folder for manuscript materials including figures and methods.
You can prepare another folder or use a diff existing one just make sure it is accessible to everyone.
AU: I made a new folder in the dropbox: Dropbox\PMC-AT Manuscripts\Manuscript 2017
Everyone, please post manuscript related materials in this folder.
==
RC: I am providing here a working template for some of the tasks.
Please post a couple of example MST figs and captions as well shortly -- I would like to check if they are on right track.
If Guan wants to add Kd standard errors later that can be done.
The working template also includes the current bibliography and below that I have excerpted some lines that need references added.
For now they can be added to bib, later these sentences with citations will be spliced back into paper.
There are questions/issues that must be addressed within the template doc.
I will add more to the template shortly.
Please post individual fiigure materials in dropbox and if you are adding figures directly to the template doc , you may repost the template doc here with your additions as well.
working template.docx
A note on another required schematic edit has been added to Fig 4 pg 4 of working template.
RC (6.7.17): On page 4 of the template above (Fig 4), in part c, the line with [A]=[A]1 should have y intercept slightly above that of the line with [A]=0 (not below as it currently is)
XG(6.7.17): Figure 4 has been modified. Please check the progress on files uploaded in dropbox/PMC AT Manuscripts\Manuscript 2017
The files are
Template Figures in progress_6.7.17
Appendix Figures in progress_6.7.17
Figure Legends in progress_6.7.17
Tables and Table Legends in progress_6.6.17
Tables of eq1 and mixed inh fittings_MnSOD.FdL_6.7.17
The following journals are currently being considered:
Biochimica et Biophysica Acta -- Proteins
Biochimica et Biophysica Acta -- General
You should review the formatting policies of this journal while working on these tasks since they will affect some of them.
I will be asking Praba to look into the journals in detail regarding esp the maximum length papers (word count, number of figs) he is able to find in the journal.
SM (5-31-2017): MST method paragraph (including description of labeling procedure)
MST method (including labeling).docx
XG(5.31.17): Figure2. 2double reciprocal mixed with respect to NAM w, wo 200 HKL; FdL; parts a,b. For FdL, choose any two experiments for inset because they are relatively similar in time series. Present single exp fit in latter case. Scale should be the same for both w,wo HKL and inset should focus on the first 30 mins
For quick look
Figure 2_2 Double reciprocal mixed_FdL.pptx
For editable version_inkscape was used.
Figure2. 2 Double reciprocal _FdL.svg
AU&XG (5.31.17): Figure 10. OAADPR inhibition Here, pairs of time series plots can be shown for comparison in each case (two figures with parts a,b in each case). Captions should be prepared.
Figure 10. OAADPr Inhibition with captions_5.31.17.pptx
For editable version_inkscape was used.
Figure10.OAADPR inhibition.svg
AU/SM/XG: 5-30: Task allocation update and some question in the document-
Task allocation-data and methods presentation assignment 5-30.docx
Task allocation-data and methods presentation assignment 5-30_RC comments.docx
RC (5/25): These tasks can be addressed by the person most familiar with each, assuming that person is free to do so. You can discuss amongst yourselves as needed for allocation of tasks. I assume you will have time during HPLC runs, e.g., to work on these.
Please review all below and let me know if some are already finished, where they can be found. Can post figs in dropbox and updates to this doc on wiki. Unless otherwise noted, assume the modulator in question is HKL. Update as tasks finish.
Note the figs and tables need to prepared in pub quality format with consistent annotations, etc, if not already done.
Some details added after initial posting. Will be updated further as needed.
data and methods presentation assignment 5-26.docx
The mst tasks refer to the mst ppt previously posted
by lab members as well as slides 5-8 in following:
cubic figure working file 5-26.pptx
The next step will be task allocation: for group members to specify who will be working on each of the tasks in the document above.
(I have suggested some allocations to lab members in latest revision above, but only for a few tasks.)
Also, to post any questions you may have. Will otherwise assume tasks are clear.
RC (6/5): See following files in PMC-AT Patents dropbox folder:
-extended_model_edits
-extended_model_modulation_edits
-free_energy_diagram_edits
for next set of edits on your figure tasks below. Also re-read the comments below, since some of the points were already mentioned.
Incorporate these fig edit tasks into your schedules asap (on Mon) and indicate there when you will be providing the next updates.
AU: 6-13-2016: I will try to finish by tommorrow.
XG(6.14.16): revised diagrams have been uploaded in dropbox/PMC-AT patents/xiangying/Figure update_6.14.16
AU: 6-14-2016: Revised pictures have been uploaded in dropbox. Folder-> Alok->Updated Figure 6-14-2016.
RC: Please straighten your arrows so the triangle is of same shape as that in original fig, and also note that the lines that you added denoting
the species name in the product release segment are not supposed to be arrows. They are just pointing to the intermediates because
it is difficult to fit them where they belong. You should remove the arrowheads and may make these lines dotted to indicate this more clearly.
Also, make the bubble in the style of that in SM's figure, starting from the middle of the original triangle rather than having two arrows pointing
toward the sides of the original triangle.
AU: 6-16-2016: I updated the figures, please see if they need edits. Thank you.
AU: Updated combined (1+2) fig.
AU: Updated combined (1+2) fig.
SM: 6-14-2016 Revised Figures for 3A_v5 (tiff and svg) have been uploaded in Dropbox/PMC-AT Patents/Sudipto/Figure Edits 6-14-2016
RC: You didn't fix the perspective as indicated in the latest sketch, in this revision.
SM: Please see updated Figures 3A_v6 (tiff and svg) below. I have also uploaded them on Dropbox.
Everyone to post fig updates to Dropbox
RC (5-25): Over the next few days, I will be assigning a few image editing and preparation tasks (and perhaps some other miscellaneous tasks) pertaining to preparation
of the next paper submission. The required files for the image prep will be in the PMC-AT patents folder in dropbox. A couple of them are already there
(extended model image drafts). Regarding the latter, instructions pertaining to the tasks have been posted on Vijayan's page and will be moved here shortly since
Vijayan may not be solely responsible to completing all these tasks. Assignments of the tasks to individual lab members will be made shortly after we have a better idea of schedules.
RC (5/27): Let me know your schedules during the next week and whether/how the tasks below may fit so I may plan.
A brief list of tasks follows. Drafts of each will be provided:
- extended sirtuin kinetic model: square diagram edits [posted to dropbox 5-24; will go in SI]
extended model 5-24.pdf
you need to make the diagrams marked "1,2,1+2, 3"
the straight arrows are annotated: k3,k4,k5,k6,koff,Pr, koff,AADPR (subject to change later)
The curvy arrows are annotated Pr, AADPR
-there are 5 intermediates along each path sometimes denoted by dots.
-a dotted line separates k6 from koff,Pr; k3-k6 correspond to "chemistry"; koff,Pr and koff,AADPR correspond to "product dissociation" stage
-you do not need to label the intermediates at this time, except for "E.AADPR.Pr" and "E.AADPR.Pr.NAM"
-diagram 1 is an expansion (zoom) of the top side/edge of the full square diagram
-diagram 2 is an expansion (zoom) of the first diagonal segment of the full square diagram
-diagram 1+2 is a combination of 1,2 that corresponds to the upper triangle in the full diagram.
the intermediates in 1,2 are connected by arrows as shown for NAM association, dissociation.
-diagram 3 is another version of diagram 1.
-a slight edit may be needed once we check whether AADPR and AcPr can co-bind (may not be possible)
RC (5/26): This task could be assigned to AU. Please indicate when you would be able to work on it vis-a-vis your schedule.
AU: 6-3-2016: Fig1: New-Fig1a-6-3-2016.svg, Fig 1 Tiff: New-Fig1a-6-3-2016.tiff
Fig 2: New-Fig2a-6-3-2016.svg, Fig 2 tiff: New-Fig2a-6-3-2016.tiff
Fig 3: New-Fig3-6-3-2016.svg, Fig 3 tiff: New-Fig3-6-3-2016.tiff
Fig 1+2 : New-Fig1-2-6-3-2016.svg, Fig 1+2 tiff: New-Fig1-2-6-3-2016.tiff
I need 1,2,1+2, and 3. In tiff or jpg format as well. As noted dropbox can be used.
AU: Fig in inkscape format. Fig1New-Fig1.svg New-Fig2.svg
SM: Figure 1 (Inkscape .svg format): Figure 1.svg and Figure 1 (tiff format): Figure 1.tif
RC: It was previously indicated by RC by email that
"1,2,3 are supposed to be insets that are not part of the original picture but rather zoom in to show certain parts of it in more detail.
This is explained on wiki and it is shown directly in the draft.
There are dotted boxes around parts of original picture (kcat arrows) and the new 1,2,3 zoom in on certain arrows.
The two kcat arrows (with their respective corners) in the original figure are expanded in this way for 1,2 respectively.
This is all shown in the draft.
Do 1,2 first then show, following which I will advise on 1+2 and 3.
The last two small arrows are koff,AADPR and koff,Pr
Before proceeding please post the responses to my wiki posting on Alok's page including the schedule of work requested."
-While the versions above may be useful, they do not adhere to the above prescriptions.It was stated that 1,2 need to be two insets that zoom into the existing sides fo the square.See the scan. You need to have a V shape that zooms in and shows the new insets outside the main fig.This is imp so we can add the names of the intermediates later.
Also, in 1 and 2, please make a version annotates E.AADPR.Pr and E.AADPR where indicated.
At this point, I want you to also start making 3 and 1+2.3 should be clear from the draft.1+2 is also drafted. Here, take 1,2 and add double sided arrows between each of the respective intermediates as shown.Since 1 starts with E.ADPR-Pr-Im and ends with E.Ac-Pr, and 2 starts with E.ADPR-Pr-Im.NAM and ends with E.Ac-Pr.NAM,1+2 will also be annotated in corresponding places with these 4 species.
Sudipto, help out if needed.
As noted figures can all be posted in PMC-AT patents dropbox if more convenient.
RC: Am waiting for the revised/add'l figures described above today.
- extended sirtuin kinetic model with modulation: cube diagram edits [posted to dropbox 5-24; will go in SI
extended model modulation.pdf
-there are 5 intermediates (same as above for a) denoted by dots
-the equilibria connecting them are labeled Kd4ii,A to Kd4v,A (I have not shown all to avoid clutter)
-you only need to show the top face of cube. As in a), you can show this as a zoom of the top face of the original cube if desired.
-you should expand the size of this image so as to avoid clutter.
RC (5/26): This task could be assigned to AU with possible help from SM. Please indicate when you would be able to work on it vis-a-vis your schedule.
SM(6/1): A first draft of Figure 3 A is attached below, in .svg format. Please advise if any modifications need to be made to this.
Figure 3 A_v1.svg
The figure in tiff format is below:
Figure 3 A_v1.tif
RC: Ok, I need you to use the zoom concept here as well for another version.
As indicated, show the original cube as is then zoom in with an inset to the top face.
Show the zoomed/expanded top face to the side.
Also, it is not clear why one of your new arrows is not labeled.
SM(6/3): Figure 3A with inset (svg format):
Figure 3 A_v3.svg
Tiff format:
Figure 3 A_v3.tif
RC: Please remove the grey diagonal arrows from the inset. Also, try to make it so the V for the inset
starts near the center of the top face of cube. And, add the dots to the kcat arrow of the inset as well.
Keep the cube otherwise exactly as it was in the original version (no new arrows i,ii,..; those only come in the inset).
You can post in dropbox if more convenient for further revisions with an indication of the filenames here.
SM(6/3): Figure 3A_v4 with inset (svg format):
Figure 3 A_v4.svg
Tiff format:
Figure 3 A_v4.tif
SM(6/13): The above two figures (Figure 3A_v4) are the latest edits. These figures have been posted on dropbox in PMC-AT Patents, subfolder: Sudipto
RC: Did you see extended model modulation edits posted on dropbox on 6/5 (as indicated at top of this wiki page). Please check that Alok has seen his
required edits as well.
SM:Below are the latest edits of Figure 3A_v5 (tiff and svg formats). I have also uploaded them on dropbox.
Figure 3 A_v5.svg
Figure 3 A_v5.tif
SM(6/15): Updated figures: Figure 3A_v6 (tiff and svg)
Figure 3 A_v6.svg
Figure 3 A_v6.tif
- sirtuin catalytic cycle free energy profile diagram preparation: to accompany cube fig.
here, free energy will be plotted against reaction coordinate [may go in main text]
various important enzyme-ligand complexes in the catalytic cycle will be plotted/labeled on the x-axis, with notations
mirroring those in the cube fig.
two profiles will be plotted: one for front face of cube, one for back face of cube (in presence of modulator).
latter will be dotted/in different color, with states annotated by".A" under the respective horizontal lines depicting the states
ideally, this fig would be made in vector graphics program (gimp) in a way that permits easy editing
(e.g., to add intermediates, modify distances between states, add annotations) during revisions.
free energy diagram draft 3.pdf
RC (5/26): This task could be assigned to XG. Please indicate when you would be able to work on it vis-a-vis your schedule.
Note: the figure is still under revision and the first draft of the vector graphics version is meant to be used only as a template for further edits/changes.
You can start by making only the profile with solid line (no "A").
"int" on x-axis can be replaced with "E.ADPR-Pr-Im" (i.e., use same terminology as in cube fig)
"coproduct" can be replaced with "E.AADPR"
"apo+OAADPR" can be replaced with "E+AADPR"
XG(5.31.2016): Two versions of free energy diagram draft3 have been prepared. The working file using inkscape was uploaded. Please make comments on that for the further modification.
Free energy diagram_working file_solid line.svg
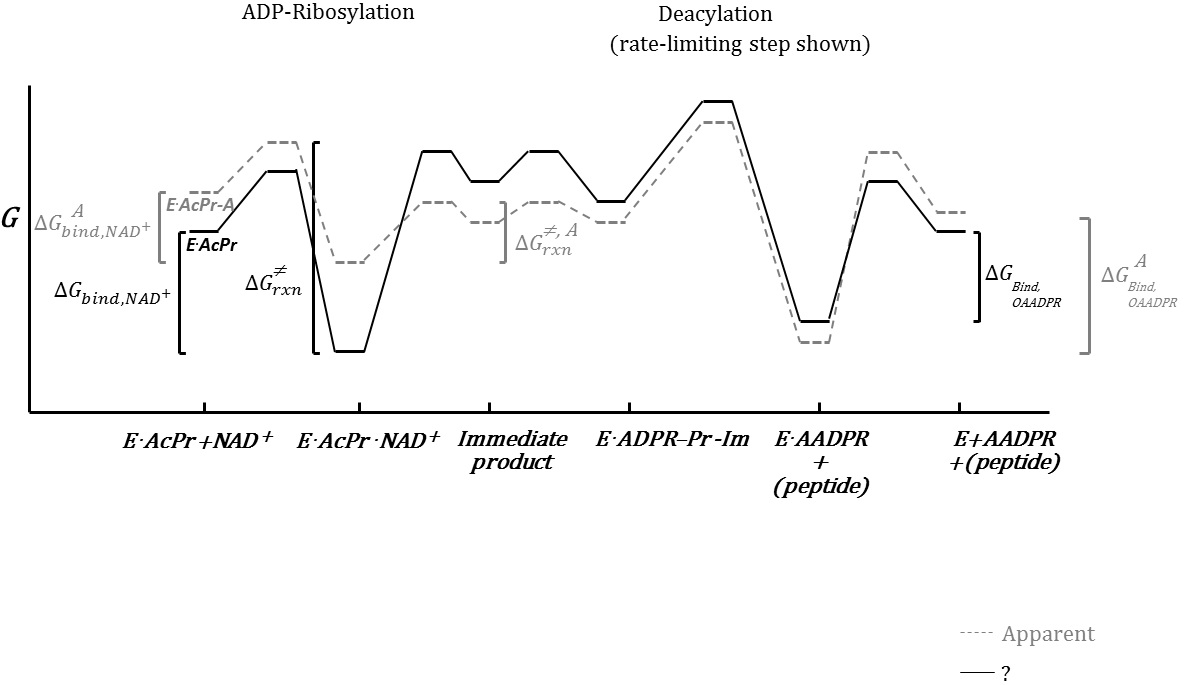
RC: The latter version is preferred. Make the following changes in the next version:
-Add right after immediate product: "E.ADPR-Pr-Im.NAM". For this, you can leave the correspond sections of the plot lines blank for now.
-You need brackets or some sort of dotted line/demarcation separating ADP-Ribosylation and Deacylation. This should occur between
ADPR-Pr-Im.NAM and E.ADPR-Pr-Im.
See draft.
-Remove the E.AcPr-A right under the horizontal lines
-Remove "O" from "OAADPR"
-"rate-limiting step shown" should be "rate-limiting chemistry step shown"
-put a gap in x-axis (and the plotted lines) a little before E.AADPR+(peptide) to shown that some of the deacylation chemistry steps are omitted
XG(6.3.16): The revision of Free energy diagram is posted below. The Inkscape working file is attached.
Free energy diagram_6.3.16.svg
RC: Typo: "Robosylation"
Immediate product and E.ADPR-Pr-Im.NAM are two different species.
As noted you don't need to specify how the line will be drawn to and from this new species for now. I will specify this later.
"A" refers to in the presence of activator (e.g., E.AcPr.NAD+.A)
Remove E.AcPr from the horizontal line in the plot.
Need delta g closure bracket in free energy fig
Deacylation bracket must start one species later
RC (6/7): I have placed a pdf with some further minor changes to the levels of states in the latest figure.
I assume you are going to put the version with ".NAM"s instead of "+NAM"s in dropbox soon as well (with these latest changes requested too).
Since these changes are subtle (and somewhat arbitrary, since we are stating the figure is not to scale), I may use the svg versions to make further level edits
going forward myself if needed.
- endnote citations and bibliography updating to add new references
RC (6/15): Once the endnote task below is done, revised protein expression/purification protocol and revised HPLC methods protocol should be posted.
SM(6/16): I have posted the revised protein expression/purification protocol for Sirt3(102-399) from AE on Task List from Sudipto wiki page. I am currently working on the FPLC protocol.
RC: A document called endnote refs doc has been posted in PMC AT Patents dropbox.
Endnote references will need to be added in the highlighted places only, for later splicing into the full paper. If you aren't sure that you have the latest bibliography, let me know.
Vijayan has a copy of the ref list of all citations previously in paper.
Once I see your schedule for completing the fig edits I will provide some more comments on where
to get some of the references here. Most of them will be obvious.
This will be started by Guan.
RC(6/6): The endnote doc in dropbox has been updated with comments on which references to use in cases where this was ambiguous.
- paper length estimate in journal format
Questions regarding the tasks can be posted here. Clarifying notes will be posted under each task above.
Minutes in response reviewer 3 and 2 comments (6-26)
Reviewer #3 question the artifact of the use of Flour-de-Lys assay.
(1)We want to emphases (1) the Flour-de-Lys method has been evaluated by other groups; and (2) the Flour-de-Lys method can be used for sirtuin inhibition studies.
(2)Make comparison of the the strengths and weaknesses of popular sirtuin activity assays
(3)Review the issue between GSK and Pfizer
(4)provide references in which Fluor-de-Lys assay and another assay were applied, and the results were in good agreement.
XG(6-26): (4)is done partially. For Reviewer 3.pptx
RC: Ok these citations make good points that we can directly quote. Can we make a case that the artifacts in some prior work were always due to interaction of fluorophore with hydrophobic moieities
on the drugs (activators), whereas we did not apply it to discover new molecules, but rather the mechanism of the C pocket binder NAM?
XG(6-27):
•Fluor-de-Lys assay used as substrates acetylated peptides conjugated to a fluorophore. It behaves as a natural SIRT1 substrates with a large hydrophobic amino acid residue [i..e, tryotophan (Trp), tyrosine (Tyr), or phenylalanine (Phe)] at positions +1 and +6.•A publication in 2008 actually showed that the use of a different acetylated peptide as SIRT1 substrate led to quite different results: in their assay system, resveratrol and fisetin showed only marginal activation of SIRT1 (about 1.3-fold), and only piceatannol had a significant activating effect (about 3-fold). The other supposed activators actually behaved as inhibitors of SIRT1: Butein, isoliquiritigenin, and quercetin reduced the activities to 0.04, 0.32, and 0.38-fold, respectively.•The so-called “artifact” is referred to the facts that “The existing STACs (identified by using FdL assay) only work with SIRT1 substrates that contain hydrophobic residues at position +1 to the acetylated lysine”. That is because they were identified via screening with a substrate that contained a hydrophobic residue mimetic–i.e., a fluorophore tag. In another word, we can call this group of STACs as “SIRT1 substrate-specific STACs”. For targeting drug design, practically, this might be a way to design/screen a candidate medicine who only targets the sirtuins for a specific disease without regulating nonspecific proteins. For example, SIRT3, as known, is a cancer suppressor and promoter. Because the substrates for this double edged sword are different, by using aformentioned method, we can design a specific STACs for the purpose of drug discovery. •Therefore, a new screen that is not biased in this way might possibly identify STACs that exhibit selectivity for SIRT1 substrates that contain other sequence signatures. It is possible that such STACs might be better therapeutics for certain aging-related diseases than the current STACs being investigated by Sirtris/GSK. •Back to our issue, like Dr Raj mentioned that the artifacts in some prior work were always due to interaction of fluorophore with hydrophobic moieities on the
drugs (activators), whereas we did not apply it to discover new molecules, but rather the mechanism of the C pocket binder NAM.For Reviewer 3_6.27.2014.pptx
Reviewer #2:
*XG will work on the Figure 2, 3.
XG(7-3): Figure2A and 2B: x-axis has been changed to same scale. To be seen clearly on the intersection of the different NAM concentrations, Figur 2C and 2D have been added. Fig_2_07.03.2014.pdf
XG(7-3): Figure 3A and 3B, the intersectionof the plots with the x-axis has been included. Fig_3_07.03.2014.pdf
*Need to emphases the Table 1, alpha value …
XG(7-3): All alphas in the manuscirpt have been highlighted in GRAY.
*provide the detailed experimental conditions on NAM inhibition of Sir2
XG(7-10): Simplified version is posted below.
------ Sinclair, D (2002) Journal of Biological Chemistry, 277: 45099-45107.Yeast Sir2p and recombinant human SIRT1 were assayed with acetylated histone substrate, dithiothreitol, and a range of NAD+ for deacetylase activity (pH=8.0) using the HDAC fluorescent activity assay/drug discovery kit (AK-500, BIOMOL Research Laboratories).
------ Sauve, AA (2003) Biochemsitry, 42: 9249-9256.Several Sir2 enzymes (Sir2Af2, ySir2, and mSir2) were assayed in the presence of KKGQSTSRHK(KAc)LMFKTEG peptide, NAD+, and selected micromolar concentrations of [carbonyl-14C]nicotinamide (pH=7.8) by HPLC for deacetylation products and NAD+.
------ Sirtris (2009) Protein Science, 18: 514-525.
Mouse SIRT33L-54-334 was assayed with AceCS2 (EILVVKRLPKTRSG-KAc-VMRRLLRKIITSEAQ), NAD+ in the presence of inhibitors (NAM and SRT1720) by mass spectrometry for the conversion of substrate to product.
XG(7-3): Two detailed experimental procedures for NAM inhibition of Sir2 are posted below. Sinclair's group used Fluor-de-Lys kit and Sauve used HPLC method.
------ Sinclair, D (2002) JBC
Recombinant glutathione S-transferase-tagged yeast Sir2p (gift of D. Moazed) and recombinant human SIRT1 (48) were assayed for deacetylase activity using the HDAC fluorescent activity assay/drug discovery kit (AK-500, BIOMOL Research Laboratories). This assay system allows detection of a fluorescent signal upon deacetylation of a histone substrate when treated with developer. Fluorescence was measured on a fluorometric reader (Cytofluor II 400 series PerSeptiveBiosystems) with excitation set at 360 nm and emission detection set at 460 nm. Reactions consisted of either 5 g of glutathione S-transferase-Sir2 or 2.5 g of SIRT1, incubated with 250 M acetylated histone substrate, 1 mMdithiothreitol, and a range of NAD concentrations as described. Reactions with the yeast and human proteins were carried out at 30 and 37 °C, respectively, for 30 min.
------ Sauve, AA (2005) Biochemsitry
Reaction mixtures of 50 mL of 50 mM potassium phosphate (pH 7.8) containing 300 mMKKGQSTSRHK(KAc)LMFKTEG peptide and 600 mMNAD+ containing selected micromolar concentrations of [carbonyl-14C]nicotinamide at 60 mCi/mmol (0, 10, 20, 30, 45, 60, 80, 90, 125, 250, 360, 600, and 1200) were reacted with 1 mMSir2 enzyme added as a 1 mL addition of concentrated enzyme. After 2 h, 10 mL aliquots were removed at 0, 30, 60, 90, and 120 min. Each aliquot was combined with 50 mL of 50 mM ammonium acetate (pH 5.0) to quench and assayed by HPLC for deacetylation products and NAD+. The chromatograms (260 nm) were obtained using 50 mM ammonium acetate (pH 5.0) as the eluanton a semipreparative Waters C-18 column (flow rate of 2.0 mL/min). Peaks for ADPR and 3’-O-acetyl-ADPR were quantified by integration. The peak for NAD+ was collected and the radiation counted. Plots of rate versus nicotinamideconcentration were fit using the curve V ) kcat[S]/([S] + Km) with the curve-fitting feature of Kaleidagraph. Plots of deacetylation rate versus nicotinamide concentration were fit to the equations described in the text. Experiments with 2 mMnicotinamide established the effects of this concentration on the deacetylation and exchange activity of the Sir2 enzyme.
*Highlight the sentences in the manuscript where reviewer #2 had issues with like
-----dixon plot, where it was mentioned in the text (YELLOW)
-----where “different substrates was used” was mentionted (BLUE)
-----where “competitive inhibition, noncompetitive inhibition” were mentioned (GREEN)
-----Where “NAM competitively inhibits mouse SIRT3 activity by Sinclair’s group” was mentioned (PINK)
XG(7-3): The aforementioned topics have been highlighted in different color (as shown above) throughout the manuscript.
PLOS ONE Manuscript_07.03.2014.docx
XG(7-8): Experimental conditions and results of Sinclair_NAM competitively inhibits mouse SIRT3 activity is listed in details.
Sinclair_NAM competitive inhibition of mSIRT3.docx
RC: After you have completed the tasks above,
- list all the points made by each referee in bullet form as was done in our other referee replies (ref comment/our reply) and list one by one all revisions we plan to make in bullet form, with references to fig numbers and page numbers (esp for referee 2).
- Leave space for statements about computational validation to be inserted by RC under referee 3.
XG(7-7): Reviewer Comments_07.07.2014.docx
- Consider adding one sentence under referee 2 that a major part of our "conclusions" pertain to the explanation for differing degrees of competitive behavior, which were also noted by Sinclair, and which constitutes at least half the paper.
XG(7-9):
- In the current manuscript, the authors reported the first time that NAM inhibition of human SIRT3 pertain some degrees of competitive behavior. which can fit into mixed noncompetitive inhibition model (
). The observed reaction rates with respect to fixed concentrations of substrate and varying concentrations of inhibitors (NAM or isoNAM) were globally fit to the aforementioned equation. The mechanisms of inhibition are determined from the alpha constant value (See Table 1). a = 1, indicates noncompetitive inhibition, a >>1, competitive inhibition, and a << 1 uncompetitive inhibition. For human SIRT3, aNAM=2.735 indicates that NAM inhibition of human SIRT3 pertain some degrees of competitive behavior.Recalled Sirtris’ 2009 work published on Protein Science, mouse SIRT3 was studied and it was found that mSIRT3 has a different mechanism of inhibition for NAM (aNAM=2.84, competitive) to those in SIRT1, SIRT2, yeast and bacterial Sir2 (noncompetitive).
- Do also consider having a para at the top for the editor indicating the contribution in terms of a generalized kinetic model for NAM inhibition of sirtuins, which does not oversimplify into noncompetitive or competitive (ref 2 may not fully understand mixed noncompetitive)
XG(7-9):
The inhibition of enzyme activity is one of the major regulatory devices of living cells, and one of the most important diagnostic procedures of the enzymologist. Inhibition studies provide important information about the specificity of an enzyme, the physical and chemical architecture of the active site, and the kinetic mechanism of the reaction. Three simple modes of enzyme inhibition are defined as follows: (A) Competitive inhibition: inhibitors bind exclusively to the free enzyme form. (B) Noncompetitive inhibition: inhibitors bind with some affinity to both the free enzyme (E) and to the enzyme-substrate complex (ES complex). When the inhibitor binds to E and ES with the same affinity, this is called pure noncompetitive inhibition. (C) Uncompetitive inhibition: inhibitors bind exclusively to the ES complex or subsequent species.
In the current manuscript, the authors have discussed a generalized kinetic model for NAM inhibition of sirtuins, which does not oversimplify into noncompetitive or competitive models, so-called mixed noncompetitive inhibition. This system may be considered a mixture of partial competitive inhibition and pure noncompetitive inhibition. When EI has a lower affinity than E for S (KS>>a KS) and ESI complex is nonproductive, the mixed noncompetitive inhibition becomes competitive inhibition. When the inhibitor binds to E and ES with the same affinity (Ki>>a Ki), the mixed noncompetitive inhibition behaves as a pure noncompetitive inhibition, where KS and aKS are the dissociation constants of the ES and ESI complex respectively (Figure below).
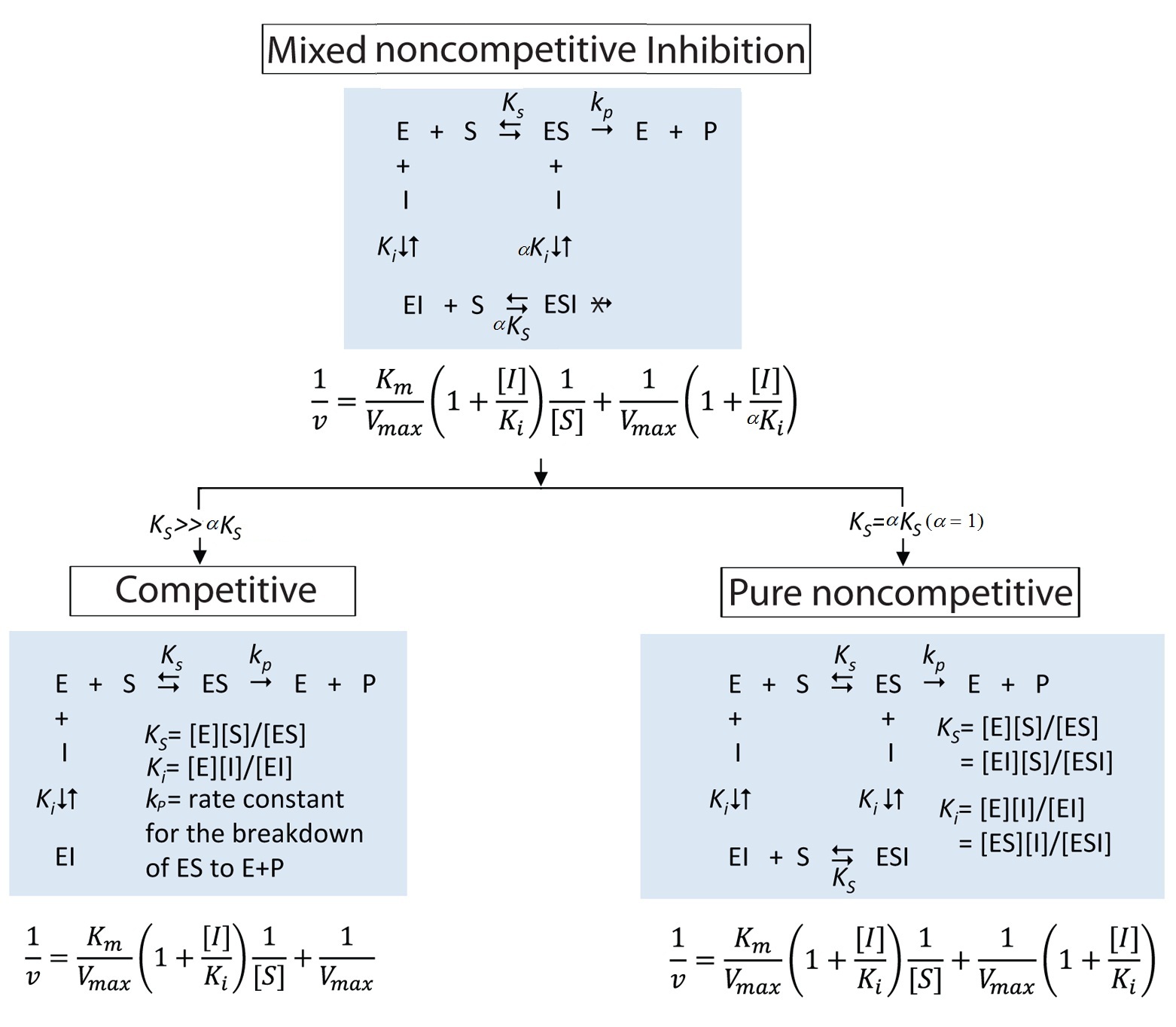
Reviewer Comments_07.09.2014.docx
You can keep doing this (in bullet form) this week if needed. After that please go back to revising 2nd paper, and then RC will write the prose English to complete the letter and will also go through the whole paper and make additional modifications to the sentences you have redlined as noted above.
RC (7-8): Some of the protocols above are too detailed to mention in this form. Is it possible to simplify and highlight the salient similarities with respect to our experiments?
What other tasks remain, if any?
After these are complete, please update on the status of the similarity search with Ping. He mentioned it in the report but some more details on how they were obtained, what/how many you are planning to buy, schedule for testing them would be useful. Did Ping rerank the compounds that ranked highly in the similarity search?
You can then return to the 2nd paper tasks.
XG(7-9): Working on simplify the aforementioned protocols. After that, will move on the "status of the similarity search".
RC (7-20): Have all referee report tasks above been completed?
XG(7-21): I am working on "add some bullet points that directly address the reviewer's point", which is the only task remain.
XG(7-17): Summary of similarity search part 1 has been attached.
Summary for Similarity Search_07.17.2014.pptx
These molecules have been sent to Ping for docking study. Ping will arrange his time since he has other tasks.
RC(7-20): Please comment on the schedule for doing these dockings.
XG(7-21): Will check with Ping for detailed schedule.
RC (7-10): I looked at the reply letter draft above. In addition to the tasks immediately above, you should try to add some bullet points that directly address the reviewer's point. For example, the protocol for Sir2 isoNAM is provided but it is not directly mentioned how this is similar to our protocol and why it addresses the reviewer's point. These can be brief bullet points, not full sentences.
Also regarding reviewer 3's comment - some bullet points about how this prior literature has validated assay can be included.
Regarding the purity of the enzyme, I assume we have no info?
XG(7-11): Regarding the purity of the enzyme, No info was provided upon the arrival of the fluor-de-Lys kit. I have emailed the Enzo technical support for details. Normally they will response within 2-3 business days.
XG(7-21): Product data sheets for SIRT1 and SIRT3 are attached below. No specific purity was mentioned.
SIRT1_human_recombinant_His-tag_BML.SE239.pdf
SIRT3_human_recombinant_His-tag_BML.SE270.pdf
XG(7-21): update reply letter draft is attached. Will go back to the 2nd paper tasks.
Reviewer Comments_07.21.2014.docx
RC (7-21): I will have a look at the above; the comments below are based on reading the previous draft (though I suspect most of the comments will still apply):
1) 1st two paras of letter - the second para is most important. Why do you refer to partial competitive but pure noncompetitive inhibition? There also needs to be some mention of the unique features of our model that do
not correspond to the pictures shown. Pictures can only be shown if there is a clear indication of how our model differs from these pictures.
2) Reviewer 2 1st point (before major comments) - should mention further details provided below. Also some grammatical errors.
3) Reviewer 2 point 1 - have other papers using this kit mentioned purity? If not this should be noted.
4) Reviewer 2 point 2 - should mention that the data in Table 1 establishes mixed noncompetitive inhibition (some explanation required).
5) Reviewer 2 point 3 - It should be mentioned that the slope of the Dixon plot indicates the degree of competitive behavior. Also, in general there needs to be indication of the page number where
we have addressed reviewer points - here the page where the Dixon discussion is provided. Some minor change may be made here explicitly indicating how the slope indicates the degree of competitive behavior.
6) Reviewer 2 point 4 - we don't seem to mention anything about isoNAM activation assay conditions. This is required to address the question.
7) Reviewer 2 minor point 1 - we don't seem to answer the question about the species origin of Sir2 Af2? Also there seems to be a typo regarding Sir3 vs Sir2. Author summary may not be considered a part of paper body.
8) Reviewer 2 point regarding URA3 - what is the conclusion here: that the sentence is ok as is? It should be indicated where we have added the original reference.
9) Have Fig/Figure been corrected?
10) Reviewer 3: a) are the papers mentioned doing both types of assays, or is the comparison done with respect to a separate paper that used Fluor-de-Lys? b) The discussion of my point near the bottom needs to be moved up, as it
is central to the argument. It should be integrated with the points regarding the artifacts, since those points alone seem to confuse the message. c) Bear in mind that the reviewer may have been referring to modulation by isoNAM (in addition
to NAM). It should be stated that computational studies demonstrate that these molecules lie in the C pocket only and hence do not have the capacity to introduce the artifacts mentioned.
11) I looked over the highlighted words in the paper and believe that, after the above changes are made, some minor edits to the language regarding competitive/noncompetitive only in the abstract and intro may be warranted. Will look into to it after 1-10 are done.
XG(7-25): Answers for above questions are in the .doc file.RC wiki 7-21_update.docx
XG(7-25): Revised figure 2 and 3 are attached.
Figure3_07.25.2014.tif